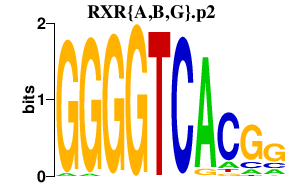
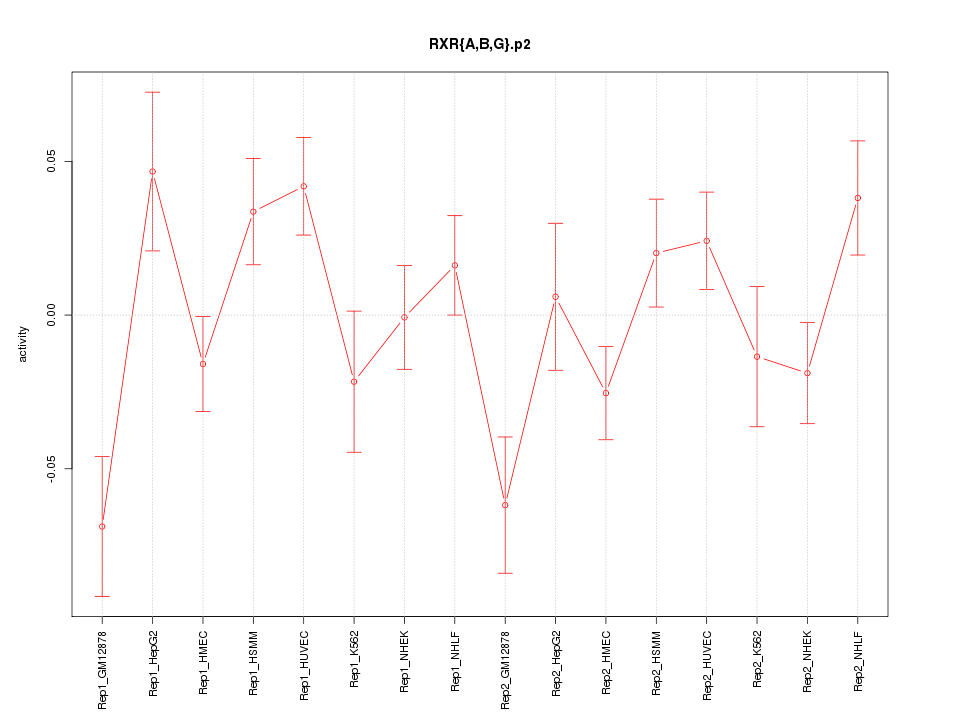
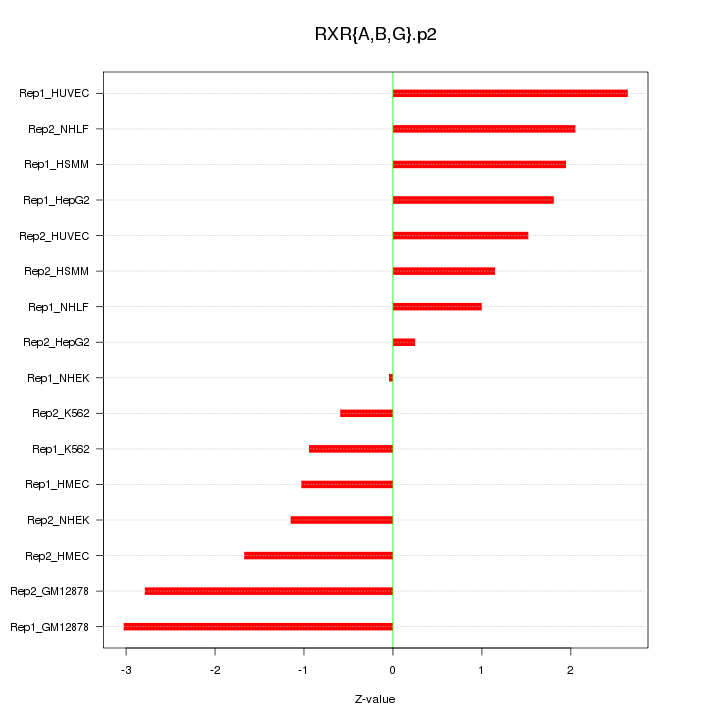
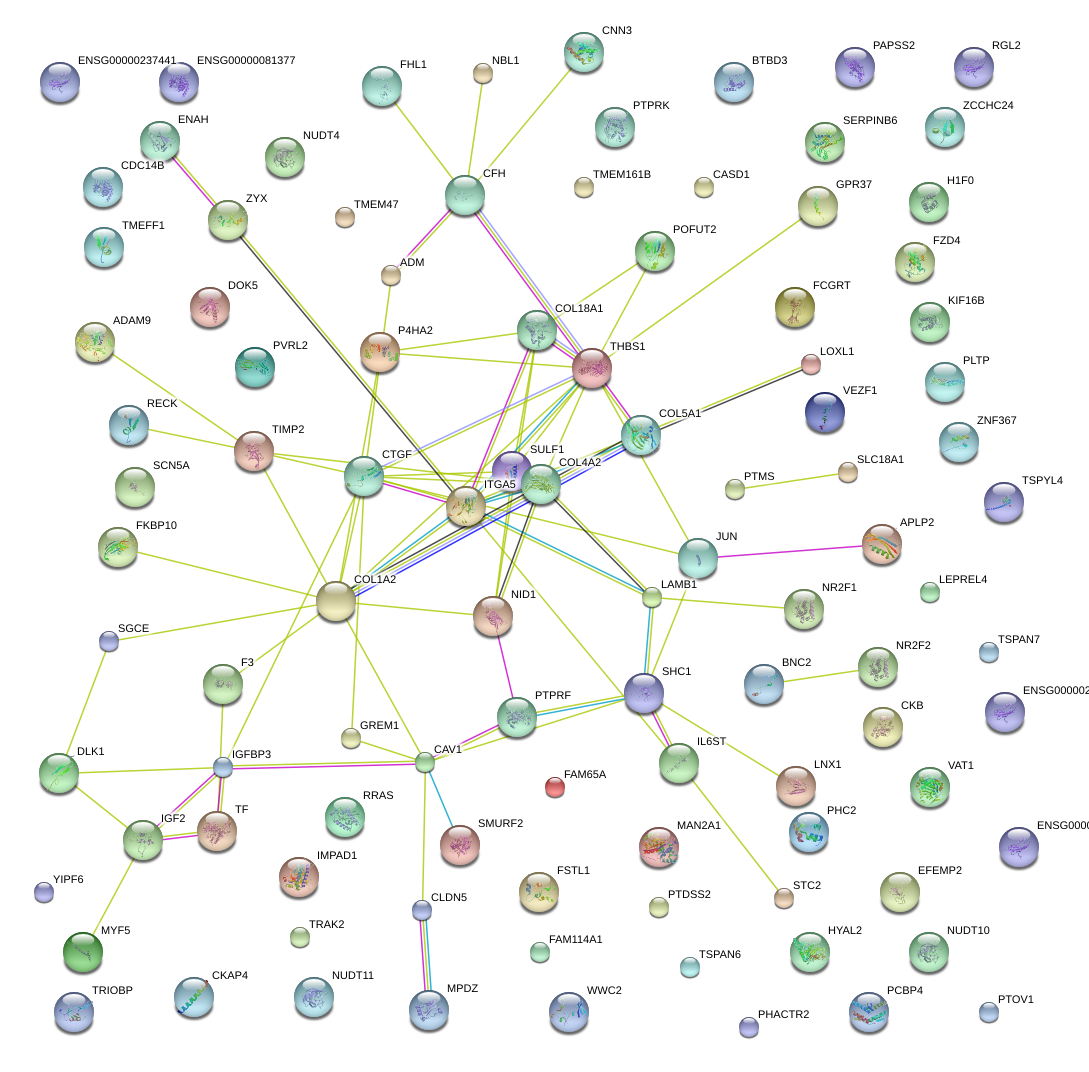
|
chr6_-_2971281
|
3.073
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr6_-_2971787
|
2.877
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr5_-_172755418
|
2.576
|
|
STC2
|
stanniocalcin 2
|
|
chr6_-_132272311
|
2.449
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr17_-_76921453
|
2.397
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr17_-_76921205
|
2.393
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr11_-_86666211
|
2.392
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr7_+_116166346
|
2.089
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_-_107643330
|
1.977
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr5_-_131563481
|
1.925
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr7_-_94285477
|
1.716
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr21_+_46825001
|
1.678
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr5_-_172755195
|
1.668
|
|
STC2
|
stanniocalcin 2
|
|
chr14_-_103988829
|
1.637
|
|
CKB
|
creatine kinase, brain
|
|
chr21_+_46825291
|
1.633
|
|
|
|
|
chr1_-_154946801
|
1.568
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr1_-_154946859
|
1.554
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_-_94285426
|
1.478
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr7_-_94285372
|
1.462
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr3_-_120169493
|
1.430
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr1_-_154946830
|
1.407
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr9_+_103235489
|
1.391
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr11_-_2162340
|
1.346
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_-_76921076
|
1.335
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr8_+_38854524
|
1.335
|
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr8_+_38854431
|
1.325
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr6_-_33266297
|
1.323
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr10_-_81205091
|
1.279
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chrX_-_99891690
|
1.279
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr7_-_45960738
|
1.277
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr15_+_33010174
|
1.213
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr9_+_36036900
|
1.192
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr11_-_65640199
|
1.172
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr19_-_50143350
|
1.152
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr11_+_10326618
|
1.130
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr15_+_74218794
|
1.130
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr6_+_143999058
|
1.126
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr15_+_74218786
|
1.124
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr22_-_19511777
|
1.104
|
|
CLDN5
|
claudin 5
|
|
chr7_+_94024201
|
1.102
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr5_+_92920592
|
1.101
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_+_137533643
|
1.090
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr15_+_74218811
|
1.083
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr9_+_103235634
|
1.065
|
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr7_+_94023872
|
1.065
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_-_34675366
|
1.057
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr1_-_225840660
|
1.051
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr3_+_133465204
|
1.043
|
|
TF
|
transferrin
|
|
chr3_+_133465250
|
1.033
|
|
TF
|
transferrin
|
|
chr3_-_50360109
|
1.026
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr1_-_95392501
|
1.026
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr17_-_62657997
|
1.023
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr3_+_133465098
|
1.022
|
|
TF
|
transferrin
|
|
chr11_+_129939787
|
1.018
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr21_-_46707748
|
1.016
|
NM_015227
NM_133635
|
POFUT2
|
protein O-fucosyltransferase 2
|
|
chr17_-_39968450
|
0.998
|
NM_006455
|
LEPREL4
|
leprecan-like 4
|
|
chr19_+_50354368
|
0.996
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr22_+_38201113
|
0.993
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr11_+_129939667
|
0.989
|
NM_001142276
NM_001142277
NM_001142278
NM_001642
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr14_-_103989195
|
0.980
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr20_+_11871364
|
0.951
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr11_+_129939807
|
0.948
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_+_143078399
|
0.936
|
|
ZYX
|
zyxin
|
|
chr19_+_45349560
|
0.932
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr1_+_43996478
|
0.928
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr6_-_2971981
|
0.928
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr4_+_38869346
|
0.928
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr3_+_133464883
|
0.911
|
NM_001063
|
TF
|
transferrin
|
|
chrX_-_51239426
|
0.903
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr11_+_129939792
|
0.903
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr10_+_89419352
|
0.899
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr11_+_129939847
|
0.891
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr12_-_54812936
|
0.887
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_106641675
|
0.882
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr14_+_101193275
|
0.881
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr8_+_70405080
|
0.871
|
|
SULF1
|
sulfatase 1
|
|
chr9_-_99329092
|
0.865
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr7_-_124405029
|
0.864
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr13_+_111138079
|
0.862
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr16_+_67571980
|
0.860
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr22_+_38092994
|
0.856
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr1_-_59248287
|
0.856
|
|
JUN
|
jun proto-oncogene
|
|
chr1_-_236228382
|
0.853
|
|
NID1
|
nidogen 1
|
|
chr4_+_184020343
|
0.846
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr17_-_56065474
|
0.843
|
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr4_-_80994183
|
0.829
|
|
|
|
|
chr1_+_182992694
|
0.825
|
|
|
|
|
chrX_+_38420690
|
0.809
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr17_+_39969200
|
0.807
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr9_-_16870664
|
0.796
|
|
BNC2
|
basonuclin 2
|
|
chr7_+_143078444
|
0.793
|
|
ZYX
|
zyxin
|
|
chr15_+_39873276
|
0.792
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr8_+_70405027
|
0.783
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr6_-_128841432
|
0.772
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr17_-_41174402
|
0.766
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr7_+_94138991
|
0.766
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr7_+_94139331
|
0.759
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr6_-_33266735
|
0.756
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr11_+_450260
|
0.753
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr1_+_19972259
|
0.750
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr7_+_143078447
|
0.749
|
|
ZYX
|
zyxin
|
|
chr5_-_131562934
|
0.747
|
NM_001017973
NM_004199
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr20_-_44539597
|
0.747
|
NM_001242921
|
PLTP
|
phospholipid transfer protein
|
|
chr6_-_33266745
|
0.746
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr17_-_41174327
|
0.744
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr11_+_450346
|
0.731
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr3_-_51993955
|
0.730
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr5_-_55290601
|
0.730
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr8_-_57906376
|
0.726
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr9_-_13279562
|
0.722
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr17_+_39969205
|
0.712
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr20_-_44539542
|
0.712
|
|
PLTP
|
phospholipid transfer protein
|
|
chr17_-_41174431
|
0.710
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr5_-_87564615
|
0.706
|
NM_153354
|
TMEM161B
|
transmembrane protein 161B
|
|
chr3_-_38691118
|
0.705
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr19_+_50016435
|
0.699
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chrX_-_51239295
|
0.699
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr8_-_57906402
|
0.693
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr2_-_202316227
|
0.683
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr6_-_116575170
|
0.682
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr1_-_33815340
|
0.677
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chrX_+_67718868
|
0.673
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr7_+_143078608
|
0.671
|
|
ZYX
|
zyxin
|
|
chr12_+_81110689
|
0.670
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr19_+_45349584
|
0.667
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr5_+_109025729
|
0.667
|
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chrX_+_135229536
|
0.667
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr20_+_53092131
|
0.661
|
NM_018431
|
DOK5
|
docking protein 5
|
|
chr6_-_33266700
|
0.658
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr1_-_94702916
|
0.657
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr20_+_35169897
|
0.657
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr15_-_90347173
|
0.654
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr1_+_51701995
|
0.654
|
|
RNF11
|
ring finger protein 11
|
|
chr19_+_50016500
|
0.652
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr17_+_39968961
|
0.651
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr19_+_50354417
|
0.650
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chrX_-_119603136
|
0.650
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr12_+_52345437
|
0.648
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr20_+_62688438
|
0.648
|
NM_198723
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr21_-_27543445
|
0.647
|
NM_001136131
|
APP
|
amyloid beta (A4) precursor protein
|
|
chrX_-_119603068
|
0.646
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr17_+_42264300
|
0.645
|
NM_001076674
NM_024107
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr15_-_90347534
|
0.640
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr19_-_36246411
|
0.637
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr4_+_88928797
|
0.636
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr7_+_143078395
|
0.631
|
|
ZYX
|
zyxin
|
|
chr14_+_24867926
|
0.629
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr21_-_46293561
|
0.627
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr21_-_46293598
|
0.627
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr17_-_40575117
|
0.627
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr21_+_35014624
|
0.625
|
NM_001001132
NM_003024
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr20_-_23066828
|
0.625
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr20_-_36793646
|
0.623
|
NM_004613
NM_198951
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr6_+_89791510
|
0.622
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr17_+_42264573
|
0.622
|
NM_177441
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr19_+_50016490
|
0.619
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr13_+_32605436
|
0.619
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr7_+_143078457
|
0.618
|
|
ZYX
|
zyxin
|
|
chr19_-_49658645
|
0.609
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr4_-_85887543
|
0.607
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_-_95392449
|
0.605
|
|
CNN3
|
calponin 3, acidic
|
|
chr15_-_48937795
|
0.593
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr11_-_27742224
|
0.591
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr15_+_96876568
|
0.589
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr13_-_96705565
|
0.588
|
NM_020121
|
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2
|
|
chr2_+_30369868
|
0.585
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr8_+_26371461
|
0.583
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr15_-_48937068
|
0.582
|
|
FBN1
|
fibrillin 1
|
|
chr13_+_110959592
|
0.580
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr10_-_15413054
|
0.579
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr1_+_19971865
|
0.577
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr10_-_33247131
|
0.576
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr2_-_113191106
|
0.576
|
NM_001164463
NM_005054
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr1_+_86046434
|
0.569
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr14_+_101193313
|
0.569
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr19_+_50353942
|
0.568
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr14_+_101193190
|
0.568
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr9_+_35792405
|
0.567
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr21_-_46293532
|
0.565
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr12_-_51420097
|
0.565
|
NM_000617
NM_001174126
NM_001174127
NM_001174128
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr9_-_116840545
|
0.564
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr20_-_25566137
|
0.559
|
NM_025176
|
NINL
|
ninein-like
|
|
chr21_-_46293609
|
0.558
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr13_-_52980234
|
0.541
|
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr1_+_182992329
|
0.541
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr3_-_55521567
|
0.538
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr22_+_44319685
|
0.535
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr17_-_36105005
|
0.532
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_28919006
|
0.531
|
NM_001193532
|
RAB42
|
RAB42, member RAS oncogene family
|
|
chr20_+_62694456
|
0.529
|
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr1_-_33815411
|
0.528
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr6_-_46459010
|
0.528
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_-_153640358
|
0.526
|
NM_001009932
NM_001009933
NM_001009934
|
DNASE1L1
|
deoxyribonuclease I-like 1
|
|
chr8_-_57906423
|
0.524
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr5_+_52776408
|
0.520
|
|
FST
|
follistatin
|
|
chr3_+_50306578
|
0.516
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr12_+_66218879
|
0.515
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr17_+_42264582
|
0.514
|
|
TMUB2
|
transmembrane and ubiquitin-like domain containing 2
|
|
chr19_+_50016606
|
0.511
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr12_-_89746244
|
0.509
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|